Call for Participants!
New Project on Organoids as NAMs for Celiac Disease Modeling
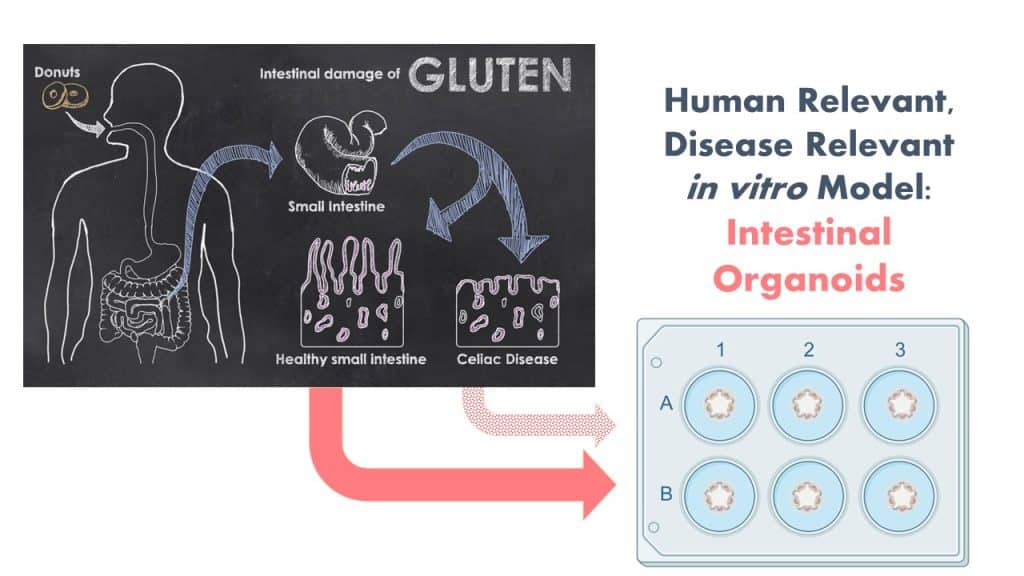
The Protein Allergens, Toxins and Bioinformatics (PATB) Committee is seeking partners to help launch a new project to advance methods for the assessment of gluten-like peptide sequences that could trigger Celiac disease (CD). The proposed pilot study will test the potential of microbial derived peptides to mimic gluten peptides using organoid models.
The project will have relevance for those involved in the evaluation of genetically modified crops, novel foods, or proteins (e.g., for food ingredients) produced via precision fermentation or microbial production systems as well as those with an interest in NAMs for food safety and clinicians/researchers involved in the study of Celiac Disease pathology and immunology.
By involving in vitro NAMs and patient derived materials to generate CD-organoids, this study is anticipated to generate both biological and methodological advancements. We are seeking additional financial support and expertise to ensure that the design and impact of the study is optimized. Your involvement will be enhanced by significant contributions in-kind by a clinical collaborator from Harvard University with expertise in developing these in vitro organoid systems! Peptides will be generated this fall and bench work to launch in early 2024.
As with all HESI projects, the success and impact of this initiative will depend on engaging thought leaders and international stakeholders, from government regulators to clinicians, to crop protection, food biotechnology, and biomedical industry experts, academics, and representatives from other relevant organizations.
Interested in finding out more?
Please contact Dr. Lucilia Mouriès at lmouries@hesiglobal.org
Project Background
Current risk assessment approaches for novel foods include amino acid sequence comparison searches, with comparisons being made to approximately 70 known 9-amino acid celiac epitopes.
However, sequence comparisons have shortcomings. Matching of novel proteins to known celiac epitopes can lead to high false positive rates; for example, many common proteins such as zeins, kafirins, and legumins as well as proteins found in pigs and cows have at least one celiac peptide match.
Furthermore, it has been proposed that changes in the Human gut microbiota can promote CD development in genetically susceptible individuals. A number of microbially derived peptides that share sequence similarities to HLA-DQ2.5-restricted gliadin determinants known to be associated with CD have been identified and shown to activate disease-relevant, gliadin-reactive T-cells isolated from CD patients. Yet, activation of T-cells in the gut is a downstream step in the cascade of events for CD. It is unknown if such peptides, when ingested as foods would pass the intestinal barrier, and reach the T-cells in the gut.
In order to reflect the clinical scenario more wholistically, we propose the use of patient-derived human gut organoids as a model system to determine the celiac potential of a protein containing a single putative CD epitope. Organoids are ideal for this use because they replicate many features of clinical disease in a disease-relevant human in vitro model system.






